One of the strengths of the ProteinTalks-DB lies in its extensive collection of mass spectrometry-based datasets from various sample types. Specifically, we offer proteomic data for 251 human tissues, encompassing 11 different types of body fluids. Additionally, we have profiled 234 brain regions in the human brain, covering both grey and white matter. For each brain sample, we provide detailed native spatial coordinates. Moreover, our database includes proteome profiling data for 45 mouse tissues.
Get started
The Search function not only enables you to look up individual genes but also allows you to conduct searches using a list of genes. You have the option to use either the gene name or the UniProt ID for your search. The names are separated by commas.

At the top of the page, you can find the UniProt entry accession, the gene and protein names, as well as indicators for their presence or detection. You can click on the icons to view the detailed expression levels in various types of tissues.
Tissues
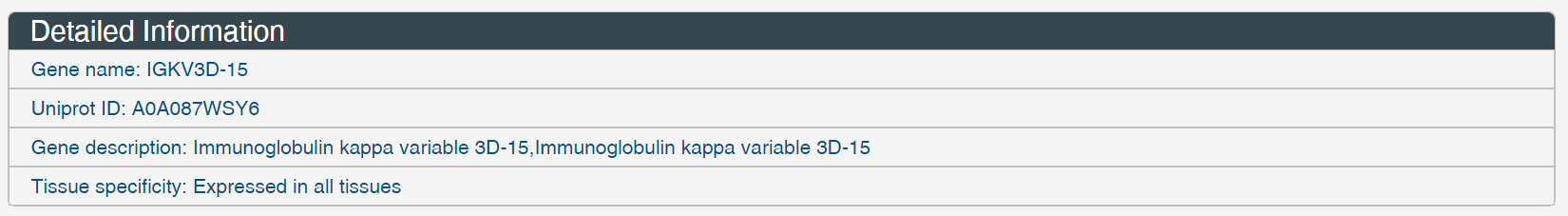
The Protein Information Card displays tissue specificity across six levels within 51 tissue types:
i. Tissue enriched
Protein levels in a particular tissue at least three times those in all other tissues
ii. Group enriched
Protein levels at least three times those in a group of 2–7 tissues
iii. Tissue enhanced
Protein levels in a particular tissue at least three times average levels in all tissues
iv. Expressed in all
Detected in all tissues
v. Not detected
Not detected in any tissue
vi. Mixed
Not included in the categories above
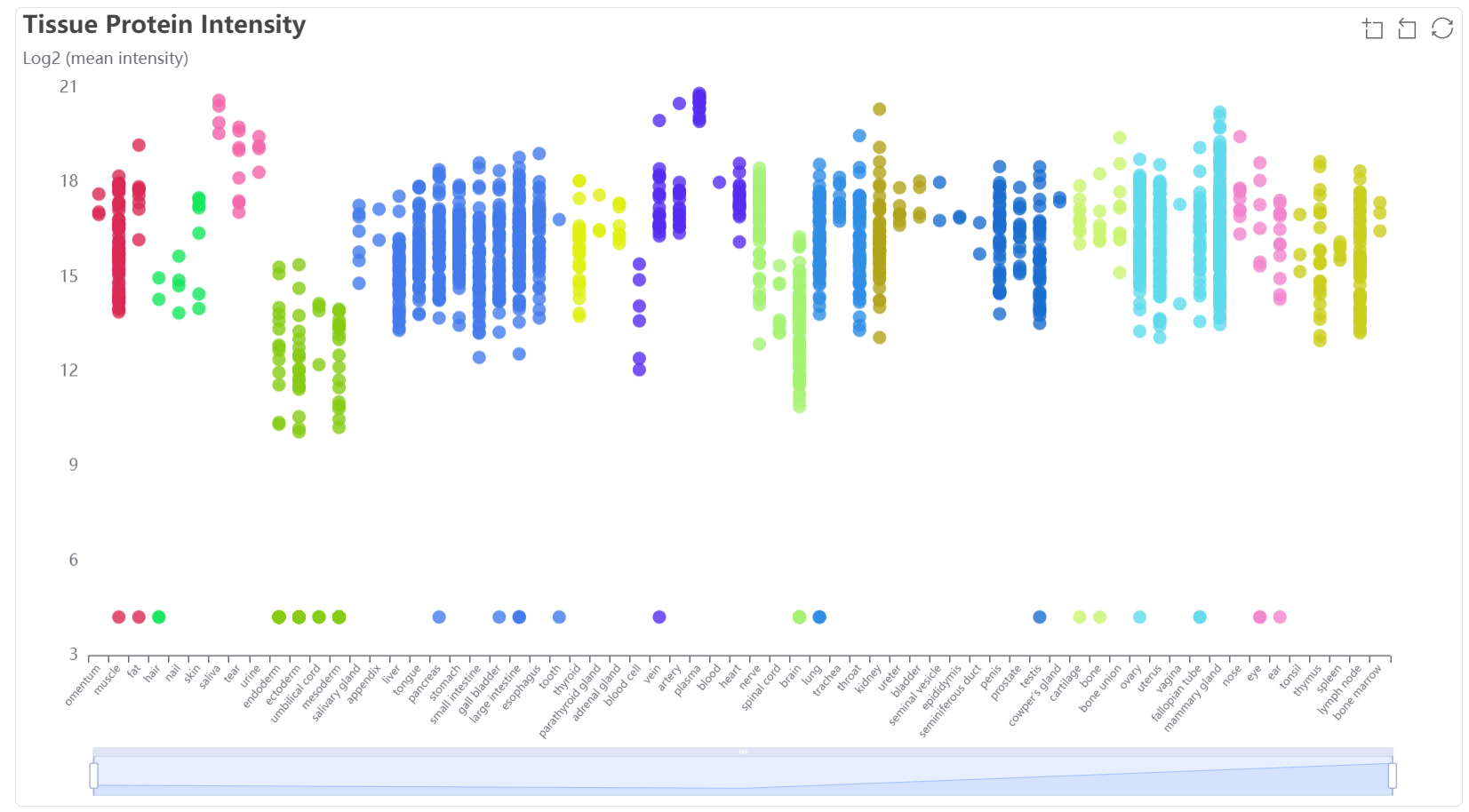
The scatter plot and bar plot show the mean expression value (log2) among major tissue types.
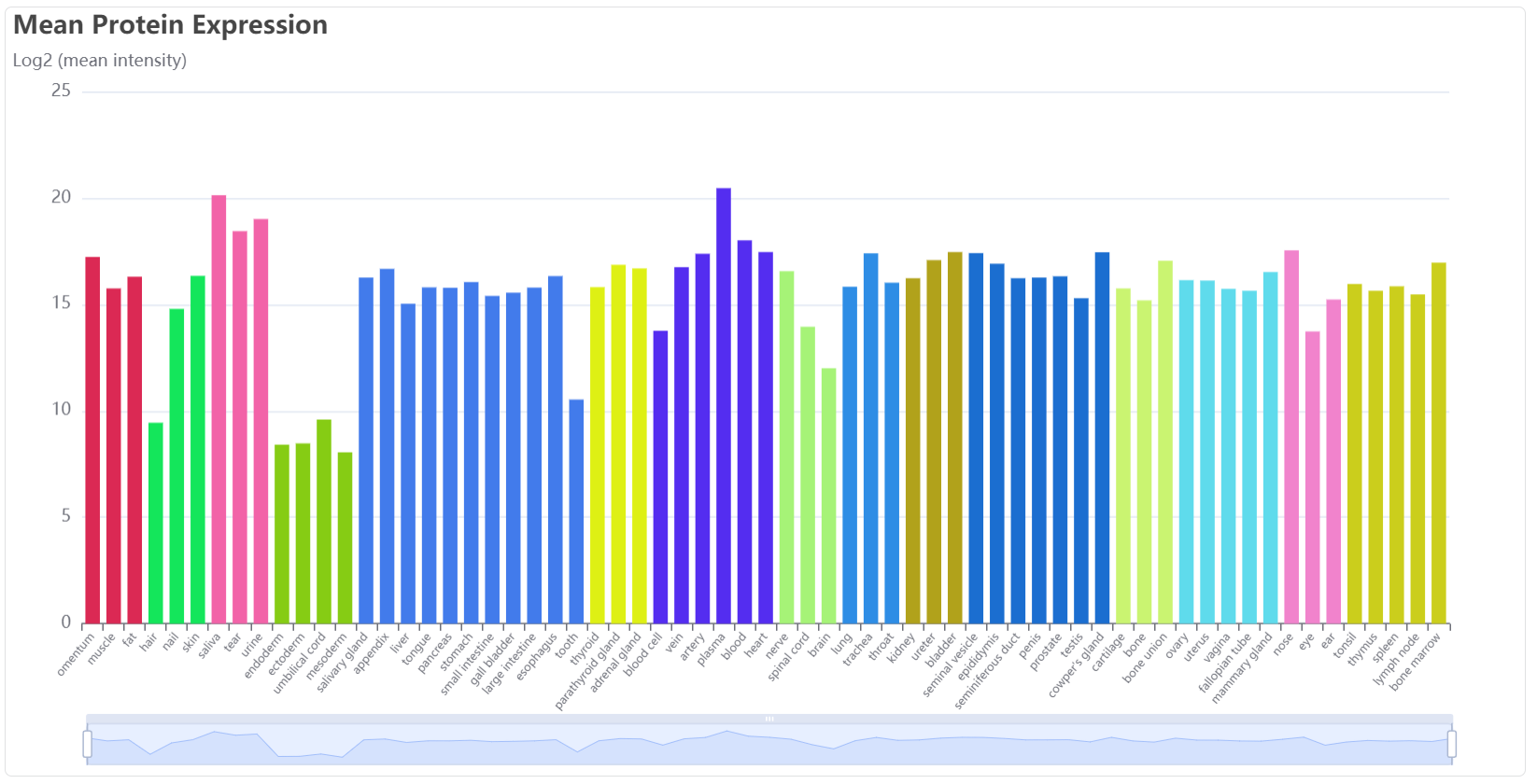
This bar plot shows the mean expression among specific tissue types. The lower pull bar allows adjustment of the displayed tissues.

Brain
Protein information card show the region specificity in grey matter and white matter. The proteins were assigned into region-specific (RS) and region-enriched (RE) using the AdaTiSS algorithm which accurately detecting tissue specificity in genes. Specifically, a protein or a transcript was considered RE protein if its standard deviation from the mean of population distribution (TS score) was greater than 2.5 in at least one brain region, while a RS a protein or a transcript was defined as having an RS score greater than 3 in a specific region and less than 2 in all other regions.

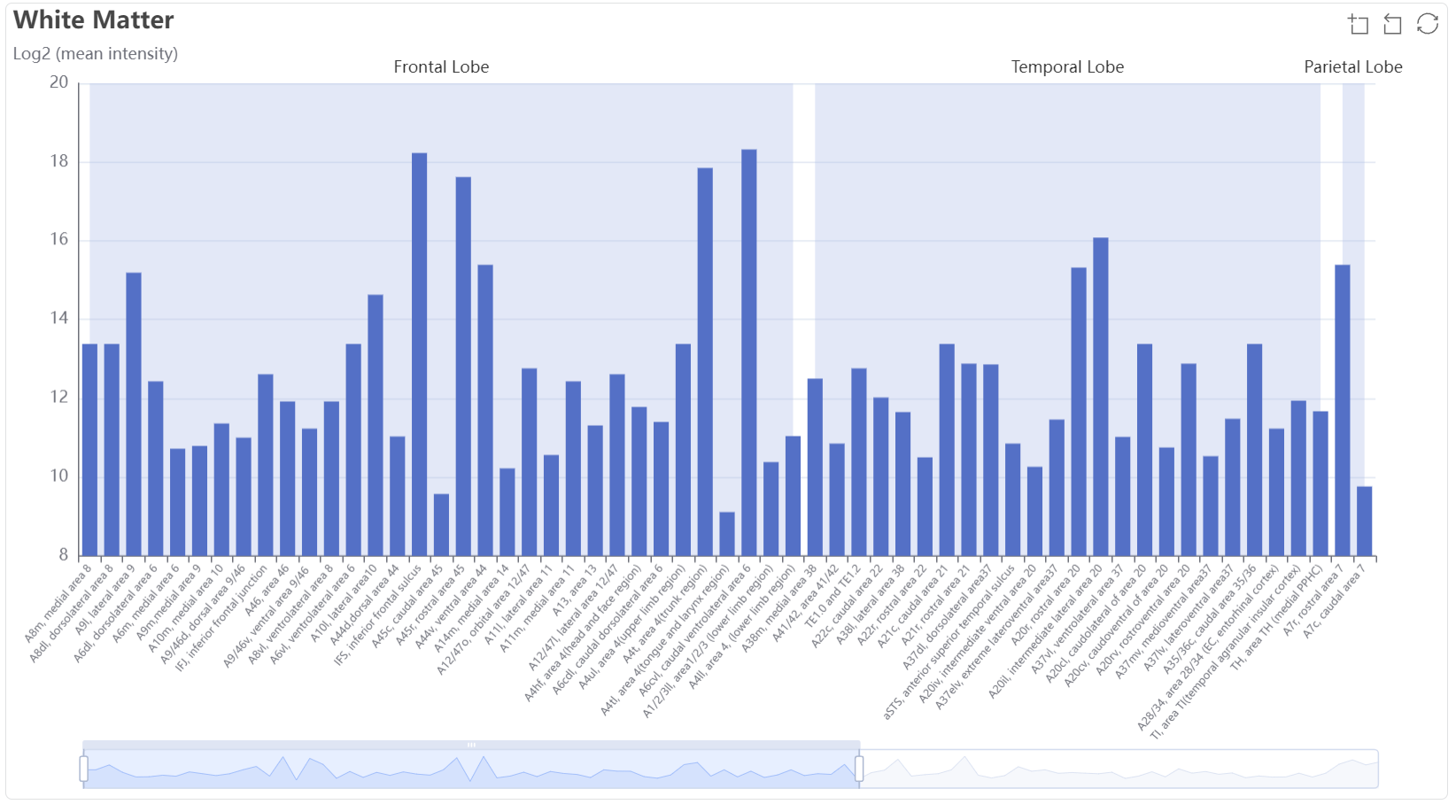
The bar plot show the mean expression value (log2) among grey matter subregions.
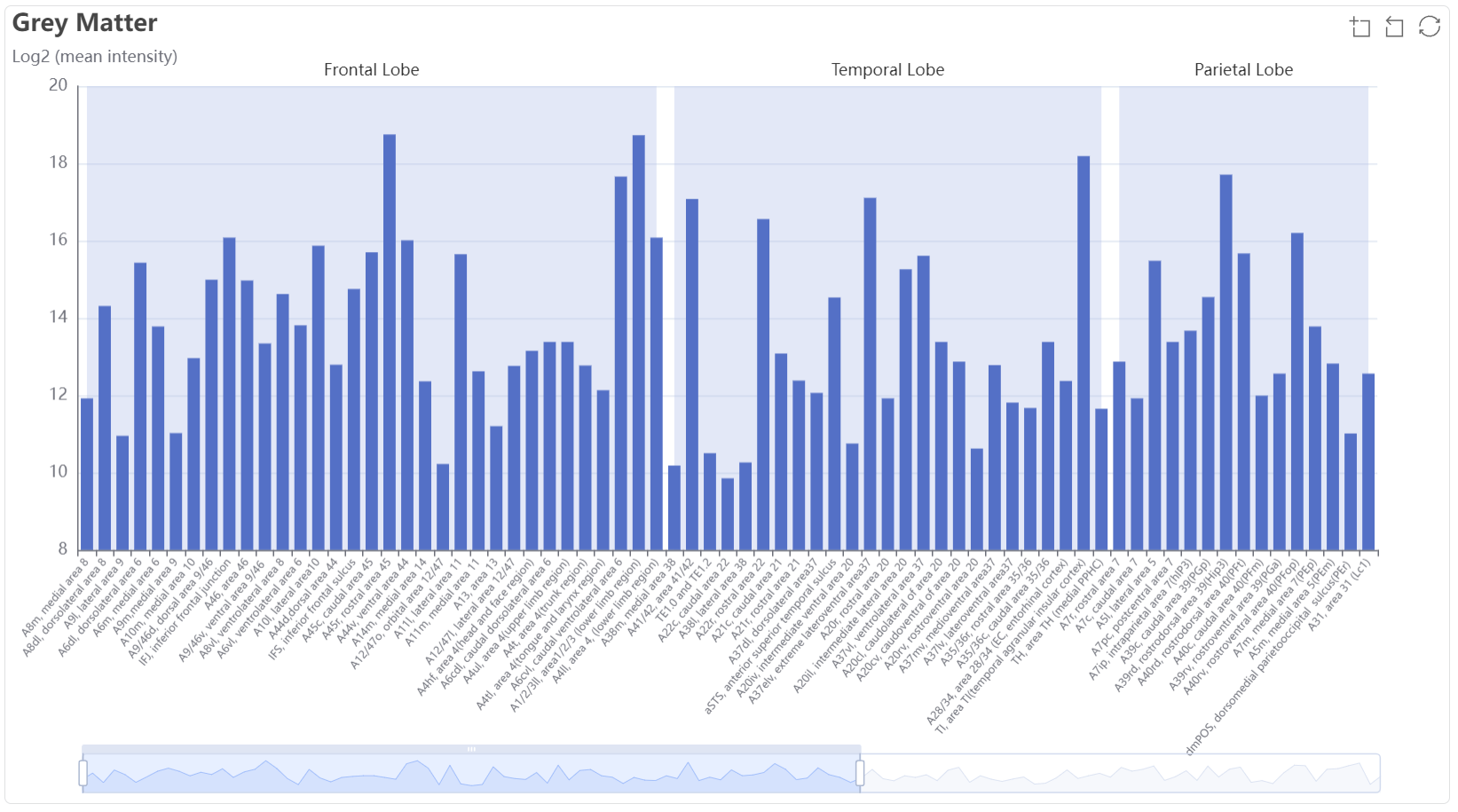
The bar plot show the mean expression value (log2) among grey white subregions.
Here we provide some details and a service for screening differential expression proteins of each dataset
Overview
-Workflow
-Differential expression proteins
Compare the proteome profiles of two selected tissue types by t-test with customized thresholds and present the differential expression proteins in a heatmap. A table of the differential expression proteins is available and downloadable.
-Functional or disease related protein lists, including:
Dysregulated proteins in paired tumors and non-tumors
For each tumor type in human tissue dataset, we compared the protein abundances in paired tumor and non-tumor samples by t-test with the thresholds of fold change and B-H adjusted p-value set at 2 and 0.05, respectively. The upregulated and downregulated proteins were available to view.
Psychological process proteins
We analyzed the proteome of hippocampus, amygdala, and 90 cortical brain regions which executing 38 psychological processes. We defined a protein as process-related for a specific psychological process if it emerges as an RE protein in over 60% of brain regions associated with that process. Our data identified 173 proteins associated with the 38 processes. Furthermore, considering the exclusive associations of anxiety with the TE1.0/2.0 region and vision color with the A7ip region in our dataset, we designated the RS proteins from these two subregions as anxiety and vision color process-related proteins. Incorporating these additions, we now possess a collective of 180 process-related proteins.
Parameters:
For differential expression proteins, we provide the following parameters to be set:
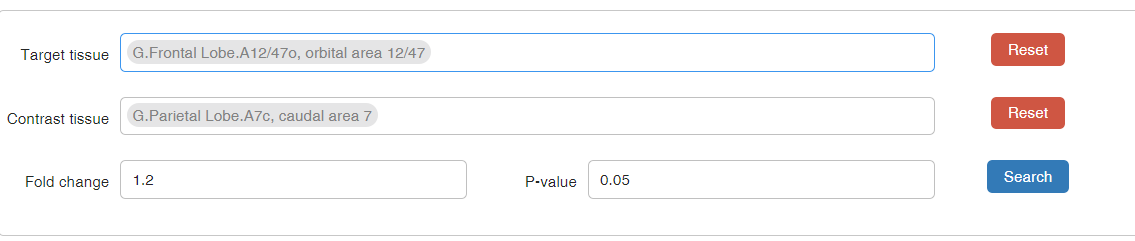
1) Target tissue: tissue name of target tissue type from the tree plot below
2) Contrast tissue: tissue name of contrast tissue type from the tree plot below
* You can select by clicking on the name on the tree.
3) Fold change: For each protein, fold change is calculated as the ratio of mean value of protein abundances in target tissues in contrast to mean value of protein abundances in contrast tissues. The threshold of fold change can be customized, usually over 1.2.
4) P-value: usually set at 0.05 for t test.
Tutorial
The module of differential expression proteins supports online comparison of the proteome of two tissue types of interest.
First, click on the selected target tissue and contrast tissue in the terminal of the circular tree plot below and the name of selected tissue type will be filled in the corresponding blank. Click on Reset to reset the selected tissue type and choose again.
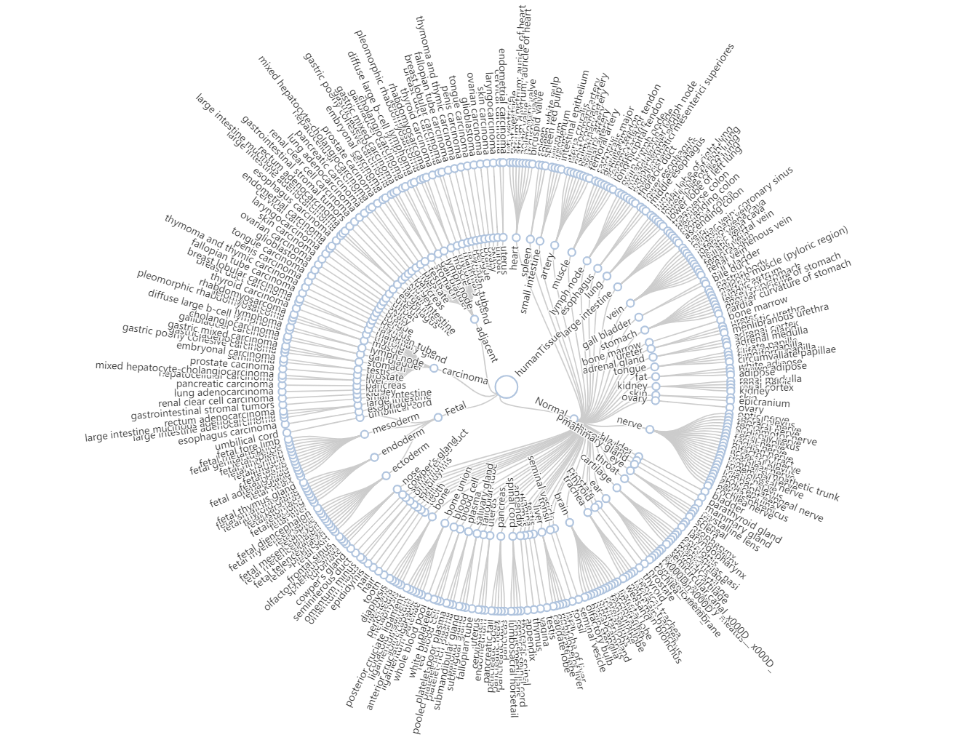
Fold change threshold: (1, Inf)
P-value threshold: 0.05
Click on Search

The differentiated expressed proteins will be presented in the form of heatmap. The rownames are the uniport ID of differentiated expressed proteins. Each column is a sample with labeled tissue type on the top of the heatmap. The webkit-scrollbar on the right of the heatmap helps to view all the proteins and zoom in. The table of the differentiated expressed proteins can be downloaded by clicking on the downloading icon
![]()